Extra-Large Area 2D/3D Imaging
Mult-Beam Scanning Electron Microscopy is an enabling technology
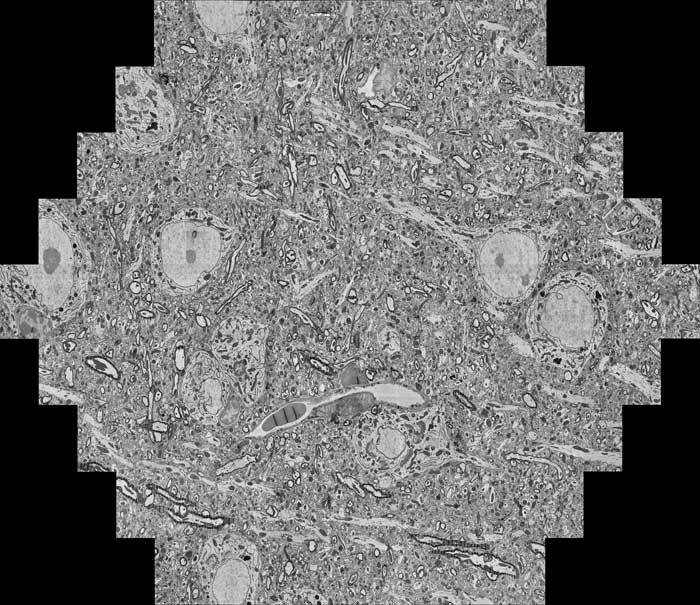
Many scientific questions need submicron image resolution over large sample areas. This is impractical with conventional SEMs (months or years acquisition time). Multi-Beam Scanning Electron Microscopes (MultiSEM) can resolve these limitations.
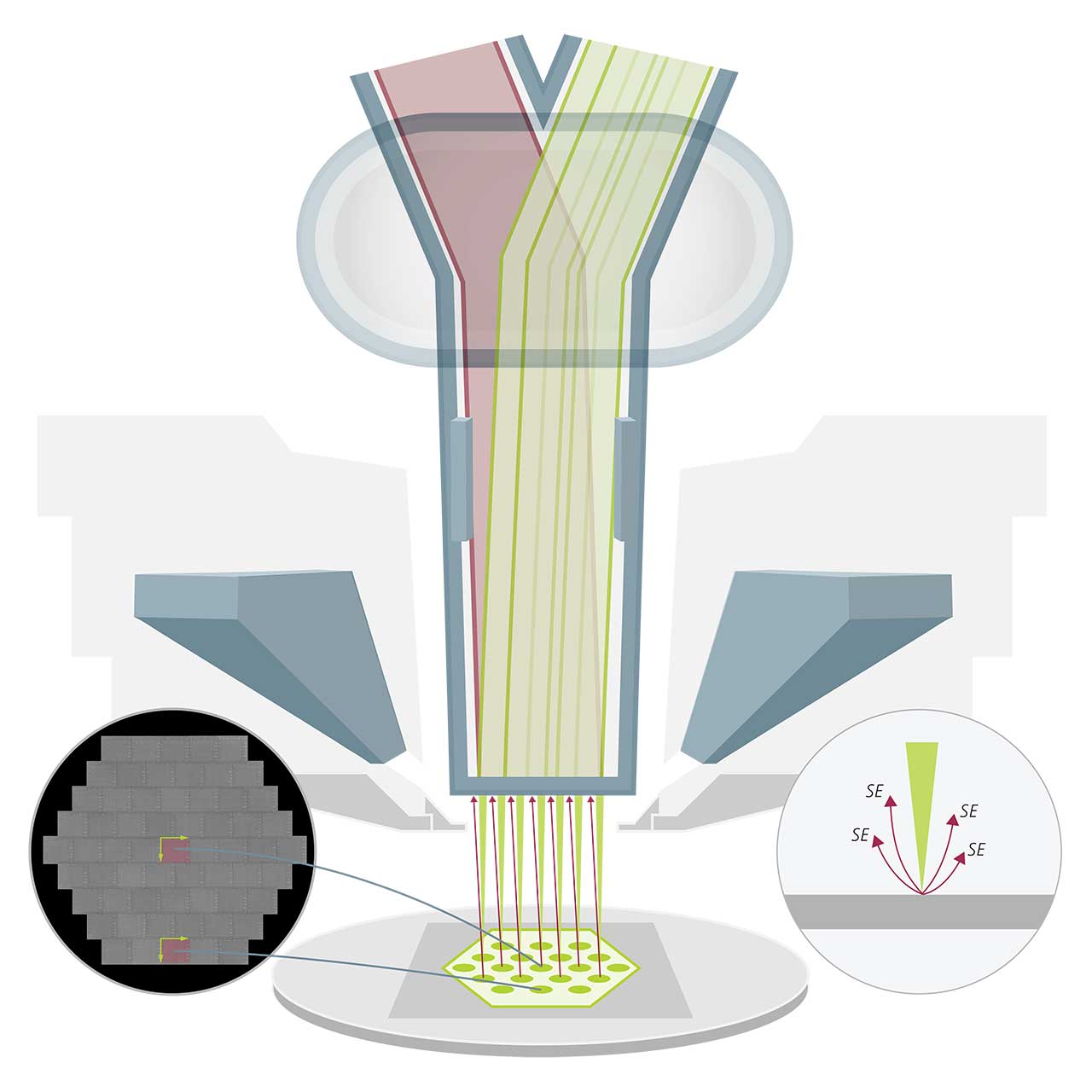
MultiSEM uses multiple electron beams (green: illumination path) and detectors in parallel. A finely tuned detection path (red) collects a large yield of secondary electrons (SE) for imaging. Each beam carries out a synchronized scanning routine at one sample position, resulting in a single sub-image. The full image is formed by merging all image tiles. A parallel computer setup is used for fast data recording, ensuring high total imaging speed. Image acquisition and workflow control are fully separated in the MultiSEM system.
More information on the instrumentation is available on the vendor websites, e.g. Carl Zeiss Microscopy (Instrumentation / Technology). NanoWorldMaps works in improving the availability of the Ultra-Fast 2D/3D Imaging technology in Europe.
Multi-beam Scanning Electron Microscopy technology
Leading the revolution in high-throughput multimodal imaging at nanometre scale
Multi-SEM is the platform technology to capture square centimetre-sized structures on a nanometer scale. It creates an ultra-dense map of the nanoworld. Artificial Intelligence (AI) methods can then be used to analyse the functionality of the sampled system or to search for and locate Points of Interest. This approach is a very good basis for multimodal analysis of the identified Points of Interest. Different types of analytical technologies such as Raman Spectroscopy (RAMAN) or Secondary Ion Mass Spectrometry (SIMS) can approach the identified points of interest and investigate them in detail.
Case studies
NanoWorldMaps members are working on a series of case studies to show the potential of Extra-large 2D/3D imaging:
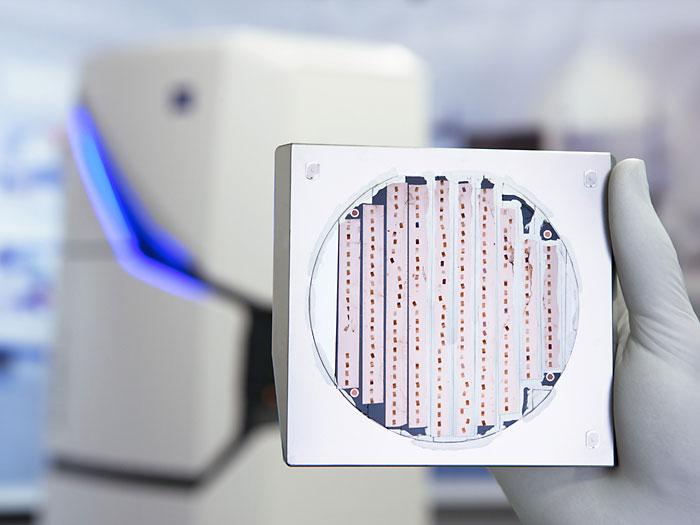
Sample preparation by microtome slicing on tape sample holders
Publications
Extra-Large 2D/3D Imaging, MultiSEM and combined multimodal approaches (RAMAN, SIMS) attracted significant scientific interest. Some publications: